% along with 10 sets of noisy letters.
numNoisy = 10;
alphabet2 = [alphabet repmat(alphabet,1,numNoisy)+randn(35,26*numNoisy)*0.2];
targets2 = [targets repmat(targets,1,numNoisy)];
net2 = train(net,alphabet2,targets2);
% ...and finally finishes.
pause % Strike any key to finish training the network...
% SET TESTING PARAMETERS
noise_range = 0:.05:.5;
max_test = 100;
network1 = [];
network2 = [];
% PERFORM THE TEST
for noiselevel = noise_range
fprintf('Testing networks with noise level of %.2f.\n',noiselevel);
Testing networks with noise level of 0.00.
errors1 = 0;
errors2 = 0;
for i=1:max_test
x = alphabet + randn(35,26)*noiselevel;
% TEST NETWORK 1
y = sim(net1,x);
yy = compet(y);
errors1 = errors1 + sum(sum(abs(yy-targets)))/2;
% TEST NETWORK 2
yn = sim(net2,x);
yyn = compet(yn);
errors2 = errors2 + sum(sum(abs(yyn-targets)))/2;
echo off
Testing networks with noise level of 0.05.
Testing networks with noise level of 0.10.
Testing networks with noise level of 0.15.
Testing networks with noise level of 0.20.
Testing networks with noise level of 0.25.
Testing networks with noise level of 0.30.
Testing networks with noise level of 0.35.
Testing networks with noise level of 0.40.
Testing networks with noise level of 0.45.
Testing networks with noise level of 0.50.
pause % Strike any key to display the test results...
% DISPLAY RESULTS
% ===============
% Here is a plot showing the percentage of errors for
% the two networks for varying levels of noise.
clf
plot(noise_range,network1*100,'--',noise_range,network2*100);
title('Percentage of Recognition Errors');
xlabel('Noise Level');
ylabel('Network 1 _ _ Network 2 ___');
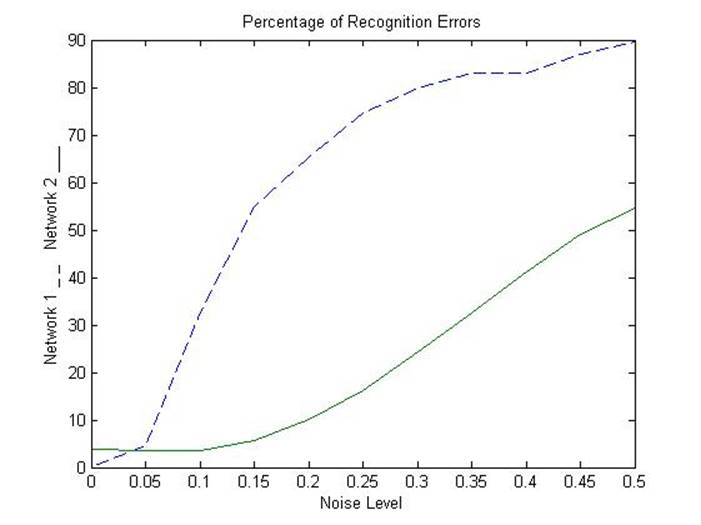
% Network 1, trained without noise, has more errors due
% to noise than does Network 2, which was trained with noise.
echo off
End of APPCR1
2.9.5. Cancer detection
Этот пример демонстрирует использование нейронной сети для обнаружения рака по данным общей спектрометрии в карте пациента. Цель – построить классификатор, который сможет различать раковых и контрольных пациентов по данным общей спектрометрии.
Методология, использовавшаяся в данной демонстрации – выбор сокращенного набора отличий или «особенностей», которые могут использоваться для определения раковых и контрольных пациентов с использованием классификатора. Такими особенностями будут уровни интенсивности ионов для определенных значений массы/заряда.
load OvarianCancerQAQCdataset.mat
whos
Name Size Bytes Class Attributes
MZ 15000x1 120000 double
Y 15000x216 25920000 double
grp 216x1 15552 cell
[feat,stat] = rankfeatures(Y,grp,'CRITERION','ttest','NUMBER',100);
P = double(Y(feat, :)); % Inputs
Cidx = strcmp('Cancer',grp); % Logical index vector for Cancer samples
T = double(Cidx)'; % Targets
rand('seed', 672880951)
net = newff(P,T,5); % create neural network
[net,tr] = train(net,P,T); % train network
testInputs = P(:,tr.testInd);
testTargets = T(:,tr.testInd);
out = round(sim(net,testInputs)); % Get response of trained network
diff = [testTargets - 2*out];
detections = length(find(diff==-1)); % cancer samples classified as cancerous
false_positives = length(find(diff==1)); % cancer samples classified as normal
true_positives = length(find(diff==0)); % normal samples classified as normal
false_alarms = length(find(diff==-2)); % normal samples classified as cancerous
Nt = size(testInputs,2); % Number of testing samples
fprintf('Total testing samples: %d\n', Nt);
%classification matrix
cm = [detections false_positives; false_alarms true_positives]
Total testing samples: 43
cm =
24 0
2 17
cm_p = (cm ./ Nt) .* 100 % classification matrix in percentages
cm_p =
55.8140 0
4.6512 39.5349
fprintf('Percentage Correct classification : %f%%\n', 100*(cm(1,1)+cm(2,2))/Nt);
fprintf('Percentage Incorrect classification : %f%%\n', 100*(cm(1,2)+cm(2,1))/Nt);
Уважаемый посетитель!
Чтобы распечатать файл, скачайте его (в формате Word).
Ссылка на скачивание - внизу страницы.